To Figure Out Which Amino Acid to Place in the Amino Acid Sequence, What Rna Molecule Is Read?

The ribosome assembles the polypeptide chain
To manufacture protein molecules, a cell must first transfer information from DNA to mRNA through the process of transcription. Then, a process called translation uses this mRNA as a template for protein associates. In fact, this catamenia of information from DNA to RNA and finally to protein is considered the central dogma of genetics, and information technology is the starting betoken for understanding the office of the genetic information in Dna.
But merely how does translation work? In other words, how does the cell read and interpret the information that is stored in DNA and carried in mRNA? The answer to this question lies in a series of circuitous mechanisms, most of which are associated with the cellular structure known as the ribosome. In society to understand these mechanisms, notwithstanding, it'southward outset necessary to take a closer look at the concept known as the genetic code.
What is the genetic lawmaking?
At its eye, the genetic code is the set of "rules" that a jail cell uses to translate the nucleotide sequence inside a molecule of mRNA. This sequence is broken into a series of three-nucleotide units known as codons (Figure 1). The three-alphabetic character nature of codons means that the four nucleotides found in mRNA — A, U, Thou, and C — can produce a total of 64 different combinations. Of these 64 codons, 61 correspond amino acids, and the remaining three represent stop signals, which trigger the end of protein synthesis. Because there are only 20 unlike amino acids but 64 possible codons, nearly amino acids are indicated past more than one codon. (Note, however, that each codon represents only i amino acid or stop codon.) This phenomenon is known as redundancy or degeneracy, and it is of import to the genetic code because it minimizes the harmful effects that incorrectly placed nucleotides can have on protein synthesis. Yet another gene that helps mitigate these potentially damaging effects is the fact that there is no overlap in the genetic code. This ways that the three nucleotides within a particular codon are a part of that codon just — thus, they are not included in either of the adjacent codons.
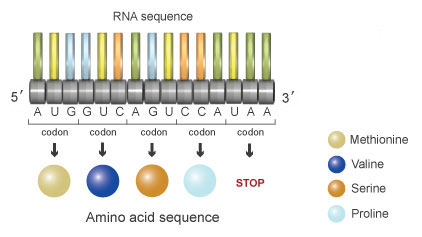
Figure 1: In mRNA, three-nucleotide units called codons dictate a particular amino acid. For instance, AUG codes for the amino acid methionine (beige).
The idea of codons was first proposed by Francis Crick and his colleagues in 1961. During that aforementioned year, Marshall Nirenberg and Heinrich Matthaei began deciphering the genetic code, and they adamant that the codon UUU specifically represented the amino acid phenylalanine. Following this discovery, Nirenberg, Philip Leder, and Har Gobind Khorana eventually identified the rest of the genetic lawmaking and fully described which codons corresponded to which amino acids.
Reading the genetic lawmaking
Back-up in the genetic code means that near amino acids are specified by more than one mRNA codon. For example, the amino acrid phenylalanine (Phe) is specified by the codons UUU and UUC, and the amino acrid leucine (Leu) is specified by the codons CUU, CUC, CUA, and CUG. Methionine is specified past the codon AUG, which is as well known as the start codon. Consequently, methionine is the outset amino acid to dock in the ribosome during the synthesis of proteins. Tryptophan is unique because it is the merely amino acrid specified past a single codon. The remaining 19 amino acids are specified past between two and half dozen codons each. The codons UAA, UAG, and UGA are the stop codons that signal the termination of translation. Figure 2 shows the 64 codon combinations and the amino acids or terminate signals they specify.
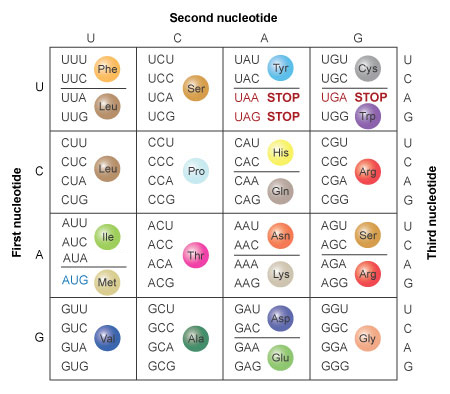
Figure ii: The amino acids specified by each mRNA codon. Multiple codons can lawmaking for the aforementioned amino acid.
What role do ribosomes play in translation?
As previously mentioned, ribosomes are the specialized cellular structures in which translation takes place. This means that ribosomes are the sites at which the genetic code is actually read by a cell. Ribosomes are themselves composed of a circuitous of proteins and specialized RNA molecules chosen ribosomal RNA (rRNA).

Figure 3: A tRNA molecule combines an anticodon sequence with an amino acid.
During translation, ribosomes move along an mRNA strand, and with the help of proteins chosen initiation factors, elongation factors, and release factors, they gather the sequence of amino acids indicated by the mRNA, thereby forming a protein. In order for this assembly to occur, however, the ribosomes must be surrounded by small but disquisitional molecules chosen transfer RNA (tRNA). Each tRNA molecule consists of ii distinct ends, ane of which binds to a specific amino acid, and the other which binds to a specific codon in the mRNA sequence because information technology carries a series of nucleotides called an anticodon (Figure three). In this mode, tRNA functions equally an adapter betwixt the genetic bulletin and the poly peptide product. (The verbal function of tRNA is explained in more depth in the following sections.)
What are the steps in translation?
Similar transcription, translation can as well exist broken into 3 singled-out phases: initiation, elongation, and termination. All three phases of translation involve the ribosome, which directs the translation process. Multiple ribosomes tin translate a single mRNA molecule at the same time, simply all of these ribosomes must begin at the kickoff codon and motion along the mRNA strand one codon at a time until reaching the finish codon. This group of ribosomes, also known as a polysome, allows for the simultaneous production of multiple strings of amino acids, called polypeptides, from one mRNA. When released, these polypeptides may be complete or, equally is often the instance, they may require farther processing to become mature proteins.
Initiation

Figure iv: During initiation, the ribosome (grey globe) docks onto the mRNA at a position about the start codon (cerise).
At the offset of the initiation stage of translation, the ribosome attaches to the mRNA strand and finds the beginning of the genetic message, called the outset codon (Figure 4). This codon is almost ever AUG, which corresponds to the amino acrid methionine. Side by side, the specific tRNA molecule that carries methionine recognizes this codon and binds to it (Figure v). At this point, the initiation stage of translation is consummate.

Figure five: To complete the initiation stage, the tRNA molecule that carries methionine recognizes the first codon and binds to information technology.
Elongation
Figure 6: Within the ribosome, multiple tRNA molecules bind to the mRNA strand in the appropriate sequence.

Figure seven: Each successive tRNA leaves backside an amino acrid that links in sequence. The resulting concatenation of amino acids emerges from the top of the ribosome.
The next step in translation, chosen elongation, begins when the ribosome shifts to the next codon on the mRNA. At this indicate, the respective tRNA binds to this codon and, for a brusque time, at that place are 2 tRNA molecules on the mRNA strand. The amino acids carried by these tRNA molecules are then bound together. Later on this binding has occurred, the ribosome shifts once more, and the first tRNA, which is no longer connected to its respective amino acid, is released (Figure vi). Now, the third codon in the mRNA strand is ready to demark with the appropriate tRNA (Figure vii). Again, the tRNA binds to the mRNA strand, the third amino acrid is added to the serial, the ribosome shifts, and the second tRNA (which no longer carries an amino acrid) is released. This process is repeated along the entire length of the mRNA, thereby elongating the polypeptide chain that is emerging from the pinnacle of the ribosome (Figure 8).

Effigy viii: The polypeptide elongates every bit the process of tRNA docking and amino acid zipper is repeated.
Termination
Eventually, later on elongation has proceeded for some time, the ribosome comes to a finish codon, which signals the finish of the genetic message. As a result, the ribosome detaches from the mRNA and releases the amino acid chain. This marks the final stage of translation, which is called termination (Figure ix).

Figure 9: The translation process terminates afterward a terminate codon signals the ribosome to autumn off the RNA.
What happens after translation?
For many proteins, translation is just the outset step in their life cycle. Moderate to extensive post-translational modification is sometimes required before a poly peptide is complete. For example, some polypeptide bondage require the add-on of other molecules earlier they are considered "finished" proteins. Nonetheless other polypeptides must have specific sections removed through a process called proteolysis. Often, this involves the excision of the outset amino acid in the concatenation (usually methionine, every bit this is the particular amino acid indicated by the beginning codon).
One time a poly peptide is complete, it has a job to perform. Some proteins are enzymes that catalyze biochemical reactions. Other proteins play roles in Dna replication and transcription. Nonetheless other proteins provide structural support for the cell, create channels through the cell membrane, or acquit out ane of many other important cellular back up functions.
Watch this video for a summary of translation in eukaryotes
montgomerywhily1994.blogspot.com
Source: https://www.nature.com/scitable/topicpage/the-information-in-dna-determines-cellular-function-6523228/
0 Response to "To Figure Out Which Amino Acid to Place in the Amino Acid Sequence, What Rna Molecule Is Read?"
Post a Comment